Lecture 2 - Filtering and Convolution¶
Announcements¶
- Please feel free to call me Scott - it has fewer syllables than "Professor" and is easier to pronounce than "Wehrwein" =)
- Office hours are posted, but also drop in anytime my door is open or email me to make an appointment.
- Project 1 is out today!
- The first (and most challenging) task is to efficiently implement convolution, which you'll be qualified to do after today's lecture.
- You'll have all the material you need by the end of tomorrow.
- I'll do a quick demo of the project in tomorrow's class.
Goals¶
Know what is meant by image noise.
Know how to filter (v) an image by cross-correlating it with a given filter (n)/kernel/weights
Know how to handle image borders when filtering:
- output sizes: full / same / valid
- out-of-bounds values: zeros, reflection, replication
Get a feel for some of the image processing operations that can and can't be accomplished using linear filtering.
- Blur, sharpen, shift
Know the properties of the convolution and cross-correlation, and some of their implications:
- linearity, associativity, commutativity
- separable filters and other compound operations
Lesson Plan¶
Revisiting image formation: image noise
Noise as motivation for neighborhood operations
Live code: mean filter
Edge handling choices:
- full, same, valid output size
- zero, reflect, repeat
Write down the discrete math definition; aside: write down the continuous definition
Live code: generalize to cross-correlation with filter weights
Mess with the filter weights:
- Horizontal blur
- Emboss
Exercise: compute a small cross-correlation result
Break
More filter weights:
- Left shift
Convolution vs cross-correlation
Properties - shift invariance; linearity; commutativity; associativity
Filter composition using associativity
Tricks using these properties:
- separable filters
- Blurring sequentially
- Sharpening
# boilerplate setup
%load_ext autoreload
%autoreload 2
import os
import sys
src_path = os.path.abspath("../src")
if (src_path not in sys.path):
sys.path.insert(0, src_path)
# Library imports
import numpy as np
import imageio.v3 as imageio
import matplotlib.pyplot as plt
import skimage as skim
import cv2
# codebase imports
import util
import filtering
The autoreload extension is already loaded. To reload it, use: %reload_ext autoreload
Let's make beans a little noisy.
Aside: different kinds of noise manifest differently. It's often helpful to have an accurate noise model for your imaging system - an mathematical understanding or approximation of the process that gives rise to noise.
A very common one is to have i.i.d. Gaussian noise. This means:
- Noise is independent for each pixel
- The mean of many samples at the same pixel will be the "true" pixel value
- The distribution around that mean will be Gaussian
# don't worry about the details here, we're just adding noise to beans
beans = imageio.imread("../data/beans.jpg")
bg = skim.color.rgb2gray(beans)
bg = skim.transform.rescale(bg, 0.25, anti_aliasing=True)
bn = bg + np.random.randn(*bg.shape) * 0.05
plt.imshow(bn, cmap="gray")
<matplotlib.image.AxesImage at 0x11a387ad0>
Brainstorm¶
If each pixel measurement is corrupted with Gaussian noise, how can we improve our guess at what the "ideal" image should have been?
Idea: mean filter
Live-pseudocode:
def mean_filter(img, filter_size):
""" Apply a square spatial mean filter with side length filter_size
to a grayscale img. Preconditions:
- img is a grayscale (2d) float image
- filter_size is odd """
H, W = img.shape
hw = filter_size // 2
out = np.zeros_like(img)
for i in range(H):
for j in range(W):
avg = 0
for fi in range(-hw, hw+1):
for fj in range(-hw, hw+1):
avg += img[i+fi, j+fj]
out[i,j] = avg / filter_size**2
# Look at mean_filter
plt.imshow(filtering.mean_filter(bn, 5), cmap="gray")
<matplotlib.image.AxesImage at 0x11aa65640>
Design Decisions - edge handling¶
(whiteboard)
Edge handling:
Output size:
- valid (only output where the whole filter overlaps the image)
- same (output same size as input)
- full (output everywhere the filter overlaps the image even a little bit)
For anything but valid, how do you handle when the filter hangs over the void?
- Zero padding
- Repeat (replicate) padding
- Reflect padding
Is a mean filter the best we can do?¶
Another example
sticks = skim.color.rgb2gray(imageio.imread("../data/sticks.png"))
plt.imshow(sticks, cmap="gray")
<matplotlib.image.AxesImage at 0x11a3aa000>
plt.imshow(filtering.mean_filter(sticks, 9), cmap="gray")
<matplotlib.image.AxesImage at 0x11a993e60>
See the lattice-like artifacts? Ick. Why is this happening?
Alternatives? *
Here's one:
plt.imshow(cv2.GaussianBlur(sticks, ksize=(9,9), sigmaX=2), cmap="gray")
<matplotlib.image.AxesImage at 0x11a9ed3d0>
Live code(?): implement filtering.filter
# test out the case of the mean filter we've been using
f = np.ones((9,9)) / (9*9)
plt.imshow(filtering.filter(sticks, f), cmap="gray")
<matplotlib.image.AxesImage at 0x11c5dab40>
# don't look behind the curtain!
g = np.zeros((9, 9))
g[4,4] = 1
g = cv2.GaussianBlur(g, ksize=(9, 9), sigmaX=2)
g /= g.sum()
plt.imshow(filtering.filter(sticks, g), cmap="gray")
<matplotlib.image.AxesImage at 0x11ad37da0>
plt.imshow(f, cmap="gray")
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x11aa65250>
plt.imshow(g, cmap="gray")
<matplotlib.image.AxesImage at 0x11c1d72f0>
How would we calculate a Gaussian kernel ourselves?¶
$$G(x, y) = \frac{1}{2 \pi \sigma^2} e^{-\frac{x^2 + y^2}{2 \sigma^2}}$$
where:
- $x, y$ are the spatial dimensions of the filter with (0,0) in the center of the kernel
- $\sigma$ is the standard deviation of the Gaussian, (determines how wide and flat vs tall and pointy it is)
Practicalities when calculating this in real life:
- You generally have to choose both a kernel size and a $\sigma$.
- For a given kernel size, you can choose sigma so it fits $\pm$ 3 standard deviations, $\sigma = (k-1)/6$
- Or for a given sigma, the kernel size that fits $\pm$ 3 standard deviations is $k = 2*\mathrm{ceil}(3*\sigma)+1$
- Not all the mass under the Gaussian fits in a finite kernel, so we usually renormalize the kernel so the weights sum to 1.
HW Problems 1 - 4¶
(1) $f \otimes w$ indicates the cross-correlation of image $f$ with filter $w$. Compute the following cross-correlation using same output size and zero padding. $$ \begin{bmatrix} 0 & 1 & 0\\ 0 & 1 & 0\\ 0 & 1 & 0 \end{bmatrix} \otimes \begin{bmatrix} 1 & 2 & 1\\ 2 & 4 & 2\\ 1 & 2 & 1 \end{bmatrix} $$
(2) Perform the same cross-correlation as above, but use repeat padding.
(3) Perform the same cross-correlation as above, but use valid output size.
(4) Describe in words the result of applying the following filter using cross-correlation. If you aren't sure, try applying it to the image above to gain intuition.
$$ \begin{bmatrix} 0 & 0 & 0\\ 0 & 0 & 1\\ 0 & 0 & 0 \end{bmatrix} $$
What else could we do with this?¶
Let's mess with filter weights to do weird stuff.
h = np.array([
[-1, -1, -1, 0, 0],
[-1, 0, 0, 0, 0],
[-1, 0, 0, 0, 1],
[0, 0, 0, 0, 1],
[0, 0, 1, 1, 1]],dtype=np.float64)
plt.imshow(filtering.filter(sticks, h), cmap="gray")
# plt.imshow(np.hstack([sticks, filtering.filter(sticks, h)]), cmap="gray")
<matplotlib.image.AxesImage at 0x11c1d4cb0>
h = np.array([
[1, 0, 0, 0, 0],
[0, 1, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 1, 0],
[0, 0, 0, 0, 1]],dtype=np.float64) / 5
plt.imshow(np.hstack([sticks, filtering.filter(sticks, h)]), cmap="gray")
<matplotlib.image.AxesImage at 0x11c5f06e0>
h = np.array([
[0, 0, 0, 0, 1],
[0, 0, 0, 1, 0],
[0, 0, 0, 0, 0],
[0, -1, 0, 0, 0],
[-1, 0, 0, 0, 0]], dtype=np.float64)
plt.imshow(filtering.filter(bg, h), cmap="gray")
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x11d9d5250>
Break¶
Math definitions¶
Discrete cross-correlation:
$$ (f \otimes g)(x, y) = \sum_{j=-\ell}^\ell \sum_{k=-\ell}^\ell f(x+j, y+k) * g(j, k) $$
Note that $g$ is defined with (0, 0) at the center.
Turns out there's a continuous version of this too! Sums become integrals:
$$ (f \otimes g)(x, y) = \int_{j=-\infty}^\infty \int_{k=-\infty}^\infty f(x+j, y+k) * g(j, k) $$ Why $\infty$? Assume zero outside the boundaries.
Properties of Cross-Correlation¶
Convolution vs cross-correlation
Properties
- Linearity: if $a$ is a scalar, $f$ is an image, and $g$, $h$ are filters, then: $$\begin{align*} a(f \otimes g) = f \otimes ag = af \otimes g \\ (f \otimes g) + (f \otimes h) = f \otimes (g + h) \end{align*}$$
- Associativity $$ f \otimes (g \otimes h) = (f \otimes g) \otimes h $$
- Commutativity? $$ f \otimes g \overset{?}{=} g \otimes f $$ Check: $$ \begin{bmatrix} 0 & 0 & 0\\ 0 & 1 & 0\\ 0 & 0 & 0 \end{bmatrix} \otimes \begin{bmatrix} 1 & 2 & 3\\ 4 & 5 & 6\\ 7 & 8 & 9 \end{bmatrix} \overset{?}{=} \begin{bmatrix} 1 & 2 & 3\\ 4 & 5 & 6\\ 7 & 8 & 9 \end{bmatrix} \otimes \begin{bmatrix} 0 & 0 & 0\\ 0 & 1 & 0\\ 0 & 0 & 0 \end{bmatrix} $$
Aside: The filter above (with just a 1 in the middle) is called the identity filter.
Convolution¶
A small modification to cross-correlation yields Convolution:
Cross-correlation ($\otimes$): $$ (f \otimes g)(x, y) = \sum_{j=-\ell}^\ell \sum_{k=-\ell}^\ell f(x+j, y+k) * g(j, k) $$ Convolution ($*$): $$ (f * g)(x, y) = \sum_{j=-\ell}^\ell \sum_{k=-\ell}^\ell f(x-j, y-k) * g(j, k) $$
This effectively flips the filter horizontally and vertically before applying it, and gains us commutativity.
orig = bg
blurred = filtering.filter(bg, g)
lost = orig - blurred
plt.imshow(orig, cmap="gray")
<matplotlib.image.AxesImage at 0x11dd8bfb0>
plt.imshow(blurred, cmap="gray")
<matplotlib.image.AxesImage at 0x11dec91c0>
plt.imshow(lost, cmap="gray")
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x11df8d790>
What if we add back what's lost?
plt.imshow(np.hstack([orig, orig + lost]), cmap="gray")
<matplotlib.image.AxesImage at 0x11deac380>
(whiteboard - equations here for posterity)
Let $I$ be the original image, $G$ be a Gaussian blur filter, and $D$ be 2 times the identity filter. Then the filtered image $I'$ is:
\begin{align*} I' &= I + (I - (I * G))\\ &= (I + I) - (I * G))\\ &= (I * D) - (I * G)\\ &= I * (D - G)\\ \end{align*}
Visual intuition:
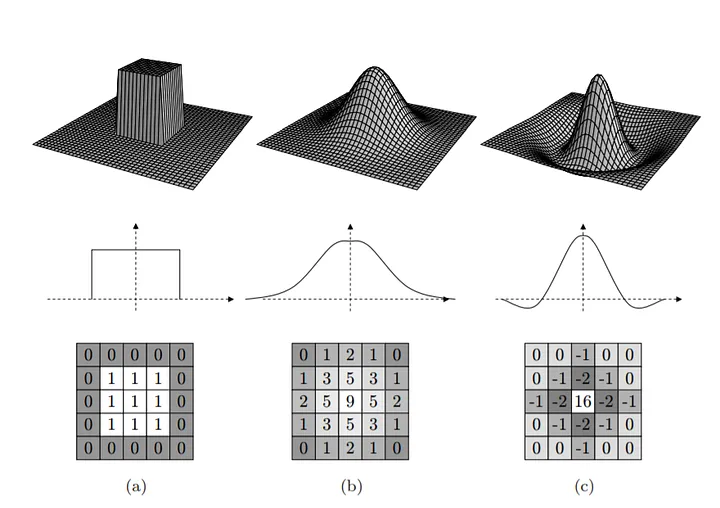
d = np.zeros_like(g)
d[4, 4] = 2
sharp = d - g
sharpened = filtering.filter(bg, sharp)
plt.imshow(np.hstack([orig, sharpened]), cmap="gray")
<matplotlib.image.AxesImage at 0x11e019640>
Efficiency trick: separable filters¶
Homework Problems 9-10:
Compute a 3x3 filter by convolving the following $1 \times 3$ filter with its transpose using full output size and zero padding: $$ \begin{bmatrix} 1 & 2 & 1\\ \end{bmatrix} $$
Suppose you have an image $F$ and you want to apply a $3 \times 3$ filter $H$ like the one above that can be written as $H = G * G^T$, where $G$ is $1 \times 3$. Which of the following will be more efficient?
- $F * (G * G^T)$
- $(F * G) * G^T$
Clarifying note: The two 1D filters being combined (e.g., $G$ and $G^T$ above) don't have to be the same filter!
For example, if
- $G = \begin{bmatrix} 1 & 2 & 1 \end{bmatrix}$
- $H = \begin{bmatrix} 1 & 0 & -1 \end{bmatrix}^T$
then
$G * H = \begin{bmatrix} 1 & 2 & 1 \\ 0 & 0 & 0 \\ -1 &-2 &-1 \end{bmatrix} $, which is a funny-looking, but quite useful filter we'll see very soon!
What can't we do with convolution?¶
Homework Problem 11:
(11) For each of the following, decide whether it's possible to design a convolution filter that performs the given operation.
Max filter: the output pixel is the maximum value among the pixels in the input window
Threshold: the output pixel is
- 255 if the input pixel is > 127
- 0 otherwise
$y$ partial derivative: the output is a finite-differences approximation of the input image's vertical derivative $\frac{\partial}{\partial y} f(x, y)$.

